Table of Contents
Introduction
FABULOUS means Fast Accurate Block Linear krylOv Solver.
BF-BCG means Breakdown Free - Block Conjugate Gradiant. BGCR means Block-Generalized Conjugate Residual. IB-BGMRES-DR means Inexact Breakdown - BlockGeneral Minimum RESidual - Deflated Restarting. IB-BGCRO-DR means Inexact Breakdown - Block-Generalized Conjugate Residual with inner Orthogonalization - Deflated Restarting.
This library implements a block BCG, BGCR, GMRes and a BGCRO linear solver for the solution of multi right-hand sides given simultaneously. Its main features are the capability to handle Inexact Breakdown and Deflated Restarting.
This work is mainly based on BlockGMRes with Inexact breakdowns and Deflated restarting, from E. Agullo, L. Giraud and Y-F Jing, SIAM Journal on Matrix Analysis and Applications 35, 4, November 2014, p. 1625–1651, [hal:hal-01067159a].
It is the combination of two former numerical schemes:
- M. Robbé and M. Sadkane, Exact and inexact breakdowns in the block GMRES method, Linear Algebra Appl., 419 (2006), pp. 265–285.
- R. B. Morgan, Restarted block GMRES with deflation of eigenvalues, Appl. Numer. Math., 54 (2005), pp. 222–236.
The BF-BCG follow the article of Hao Ji & Yaohang Li
- A breakdown-free block conjugate gradient method, BIT Numer Math (2017) 57:379–403, DOI 10.1007/s10543-016-0631-z, Springer Science+Business Media Dordrecht 2016
The IB-BGCRO-DR algorithm it is based on works of L. Giraud, Y.-F. Jing, Y.-F. Xiang
- A block minimum residual norm subspace solver for sequences of multiple left and right-hand side linear systems. [Research Report] RR-9393, Inria Bordeaux Sud-Ouest. 2021, pp.60. hal-03146213v2
And was developed in accordance of the Lyncs PARCE-6IP-WP8 project.
External links
IB-BGMRes-DR article from SIAM
Description
The core of the library is sequential while the matrix-vector product and preconditioner kernels are provided by the user as callbacks (see below). It implements a classical BGMRes (Block GMRes), a BGMRes with Inexact Breakdowns and a BGCRO with Inexact Breakdowns . It relies on Lapack and Blas kernels for the computational part. A C Api is available (see fabulous_basic.h). Because the stopping criterion is based on a backward error, only right-preconditioner is supported that does not impact this criterion.
This library does not implement the Matrix Product. Thus, the user must provide to the library a Matrix x Block of vector product. To provide it, the user must pass it through a function pointer with the C Api, or by providing a Matrix class similar to the one displayed and used inside the examples provided. (testMatrixMarket ...). Similarly, the user can provide a right preconditionner, to be used during computation.
Features
The current state of the library provides:
- BCG and BF-BCG implementation
- BCGR implementation
- Standard Block GMRes implementation
- Inexact Breakdown detection during Arnoldi's procedure, and during restart
- Deflated Restart
- IB-BGCRO-DR implementation
- Multi precision: the user can provide a targeted tolerance for each vector in the right hand side block, or a common one.
- Four different orthogonalization scheme: Classical Gram Schmidt, Modified Gram Schmidt, and their iterated counterpart.
Performances
The graphs below illustrate the numerical behaviour and feature of the block solver. The graph on top displays the envelop of the convergence history (i.e., the maximum and minimum of the backward error among all the right-hand sides) as a function of the number of matrix-vector products performed. The graph on the bottom shows the numerical feature, the vertical bars denote the size of the block used for each search space expansion. It can be seen that the inexact breakdown mechanism enables to reduce the block size when the convergence takes place. For this experiment, the maximal dimension of the search space was 350, which implies a restart when this dimension is reached. The restart are characteristed on the time curve by a change of the slop related to the lower cost of the orthogonalisation.
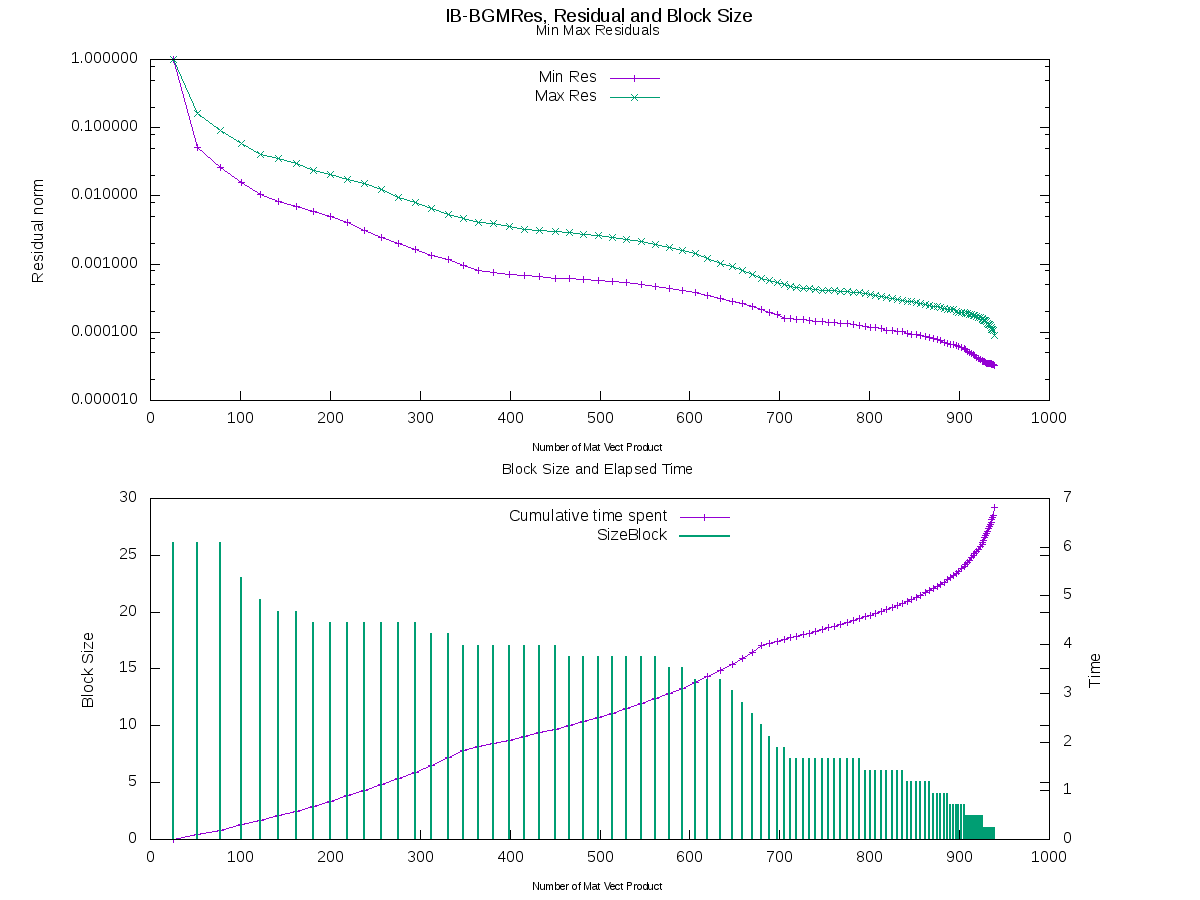
Comparaison of available algorithms
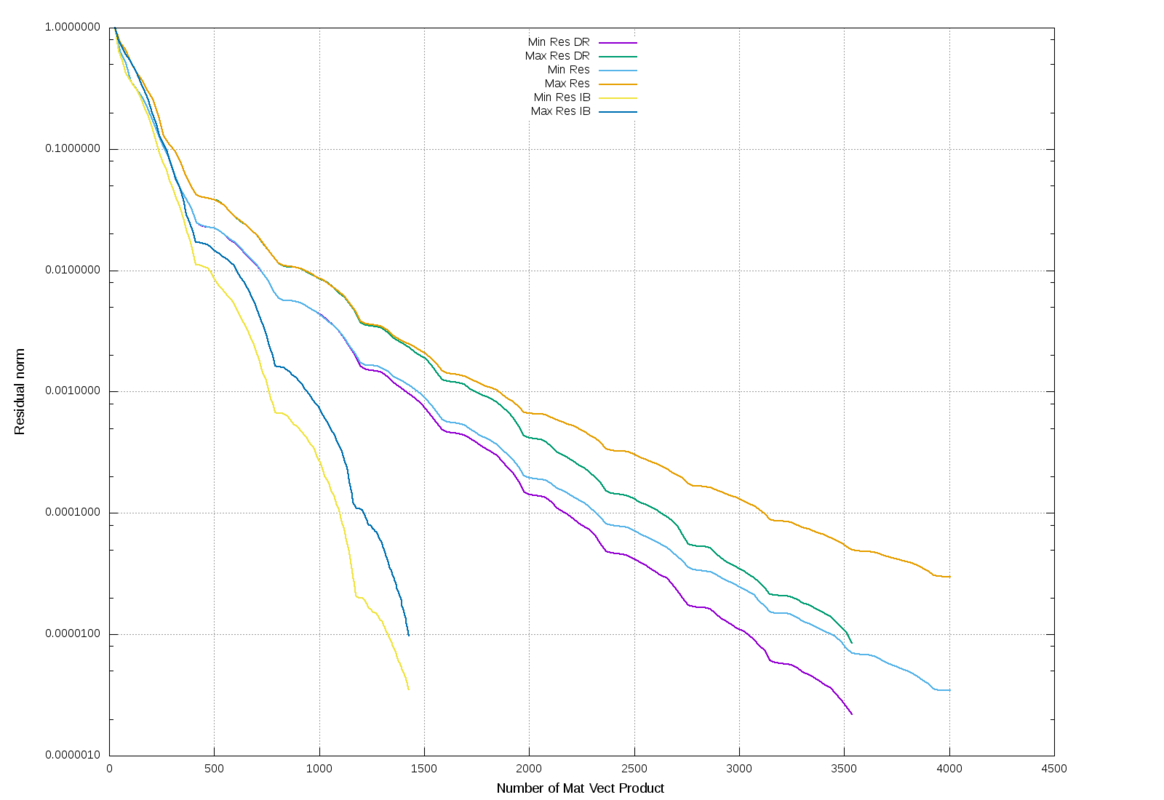
The following graph display the min and max residual norm during execution for 3 versions: BGMRES, IB-BGMRES and BGMRES-DR.
The matrix is the same as above, the maximum size for the Krylov space is set to 400, there are 26 RHS vectors.
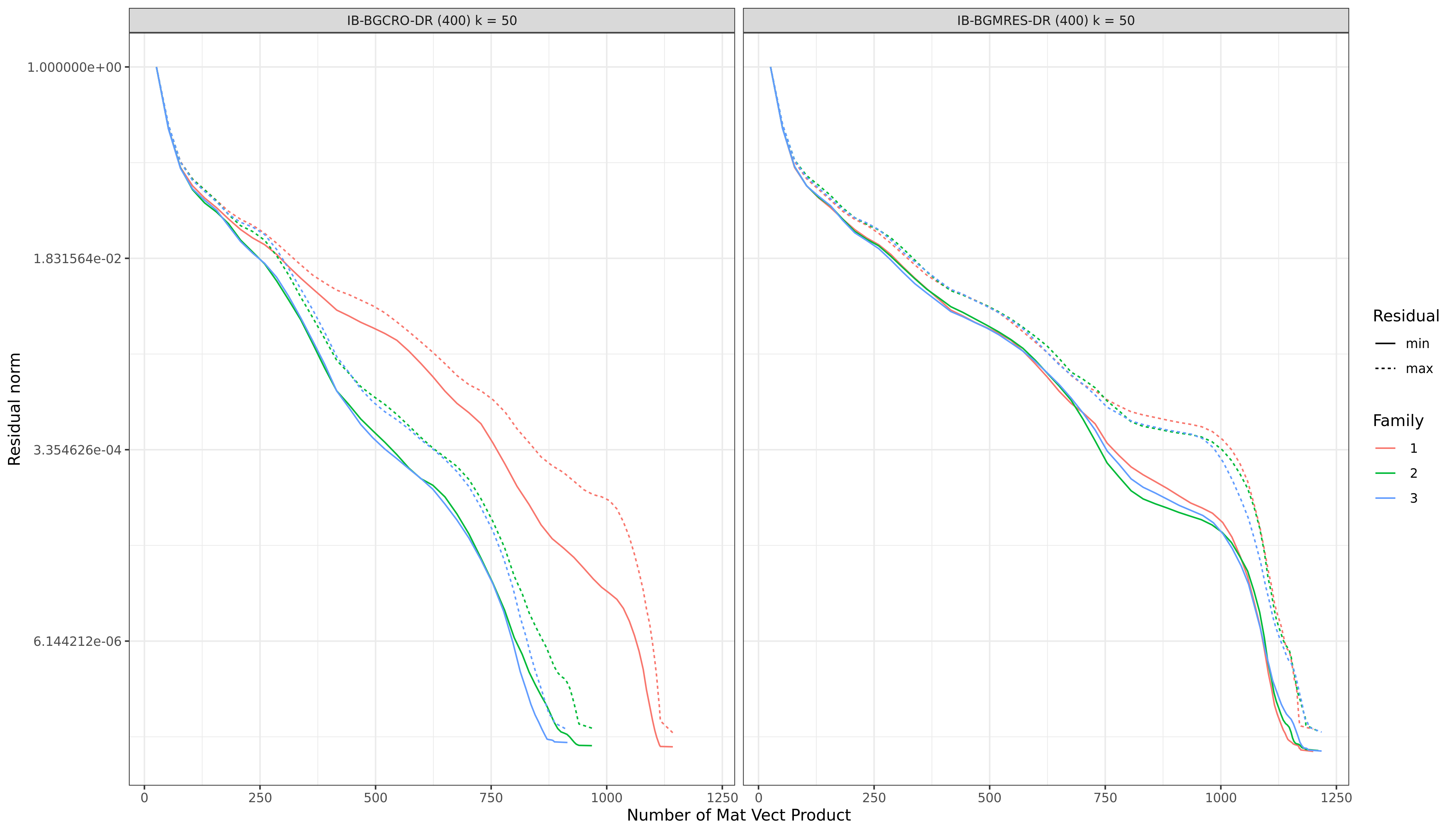
The graph below shows the main diffrence between IB-BGMRes-DR and IB-BGCRO-DR. The left part of the graph shows the speedup effect of IB-BGCRO-DR's recycling policy on 3 concecutive blocks (families) of 26 right-hand-side each. The right part of the graph correspond to IB-BGMRes-DR called on the same 3 concecutive blocks. The maximum size for the Krylov space is set to 400, and the deflation space is of size 50.
Quick Start
Prerequisites
Required:
- git.
- cmake.
- c/c++ compiler supporting c++17 standard.
- Lapacke and cblas implementation.
How to build
How to build the library:
- Create a build directory: mkdir Build
- cd inside directory cd Build
- Run cmake cmake ..
- Run ccmake to configure if needed ccmake ..
- Compile make && make install
CMake linking
The cmake has been updated to be more modern, the FindChameleon.cmake file is no longer requiered, FabulousConfig.cmake will automaticaly find dependencies.
To link the Fabulous to your own library please use the following corresponding target.
- FABULOUS::fabulous_cpp
- FABULOUS::fabulous_c_api
- FABULOUS::fabulous_fortran_api
For previous Fabulous-Chameleon Users
Chameleon is deprecated in this version, please checkout Fabulous-1.0.1 to have the latest Chameleon supported version.
With the modern CMake, use CHAMELEON_ROOT=$PATH_CHAMELEON or CMAKE_PREFIX_PATH="$PATH1:$PATH2" to indicate the location of the Chameleon.
Contacts
IB-BGMRes-DR / IB-BGCRO-DR are currently under development by the HiePACS team (https://team.inria.fr/hiepacs/).
Authors
- Luc Giraud (luc.giraud at inria.fr)
- Cyrille Piacibello (cyrille.piacibello at inria.fr)
- Thomas Mijieux (thomas.mijieux at inria.fr)
- Matthieu Simonin (matthieu.a.simonin at inria.fr)
